#include<Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector add1(NumericVector x) {
NumericVector ans(x.size());
for (int i = 0; i < x.size(); ++i)
ans[i] = x[i] + 1;
return ans;
}7 Rcpp
When parallel computing is not enough, you can boost your R code using a lower-level programming language1 like C++, C, or Fortran. With R itself written in C, it provides access points (APIs) to connect C++/C/Fortran functions to R. Although not impossible, using lower-level languages to enhance R can be cumbersome; Rcpp (Eddelbuettel and François 2011; Eddelbuettel 2013; Eddelbuettel and Balamuta 2018) can make things very easy. This chapter shows you how to use Rcpp–the most popular way to connect C++ with R–to accelerate your R code.
7.1 Before we start
You need to have Rcpp installed in your system:
install.packages("Rcpp")You need to have a compiler
And that’s it!
7.2 R is great, but…
The problem:
As we saw, R is very fast… once vectorized
What to do if your model cannot be vectorized?
The solution: Use C/C++/Fotran! It works with R!
The problem to the solution: What R user knows any of those!?
R has had an API (application programming interface) for integrating C/C++ code with R for a long time.
Unfortunately, it is not very straightforward
7.3 Enter Rcpp
One of the most important R packages on CRAN.
As of January 22, 2023, about 50% of CRAN packages depend on it (directly or not).
From the package description:
The ‘Rcpp’ package provides R functions as well as C++ classes which offer a seamless integration of R and C++
7.4 Why bother?
To draw ten numbers from a normal distribution with sd = 100.0 using R C API:
SEXP stats = PROTECT(R_FindNamespace(mkString("stats"))); SEXP rnorm = PROTECT(findVarInFrame(stats, install("rnorm"))); SEXP call = PROTECT( LCONS( rnorm, CONS(ScalarInteger(10), CONS(ScalarReal(100.0), R_NilValue)))); SET_TAG(CDDR(call),install("sd")); SEXP res = PROTECT(eval(call, R_GlobalEnv)); UNPROTECT(4); return res;Using Rcpp:
Environment stats("package:stats"); Function rnorm = stats["rnorm"]; return rnorm(10, Named("sd", 100.0));
7.5 Example 1: Looping over a vector
add1(1:10) [1] 2 3 4 5 6 7 8 9 10 11Make it sweeter by adding some “sugar” (the Rcpp kind)
#include<Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector add1Cpp(NumericVector x) {
return x + 1;
}add1Cpp(1:10) [1] 2 3 4 5 6 7 8 9 10 117.6 How much fast?
Compared to this:
add1R <- function(x) {
for (i in 1:length(x))
x[i] <- x[i] + 1
x
}
microbenchmark::microbenchmark(add1R(1:1000), add1Cpp(1:1000))Unit: microseconds
expr min lq mean median uq max neval
add1R(1:1000) 57.507 57.9885 80.43857 58.3585 60.5535 2124.504 100
add1Cpp(1:1000) 2.865 3.1860 14.13444 3.4470 8.7820 900.510 1007.7 Main differences between R and C++
One is compiled, and the other interpreted
Indexing objects: In C++ the indices range from 0 to
(n - 1), whereas in R is from 1 ton.All expressions end with a
;(optional in R).In C++ object need to be declared, in R not (dynamic).
7.8 C++/Rcpp fundamentals: Types
Besides C-like data types (double, int, char, and bool), we can use the following types of objects with Rcpp:
Matrices:
NumericMatrix,IntegerMatrix,LogicalMatrix,CharacterMatrixVectors:
NumericVector,IntegerVector,LogicalVector,CharacterVectorAnd more!:
DataFrame,List,Function,Environment
7.9 Parts of “an Rcpp program”
#include<Rcpp.h>
using namespace Rcpp
// [[Rcpp::export]]
NumericVector add1(NumericVector x) {
NumericVector ans(x.size());
for (int i = 0; i < x.size(); ++i)
ans[i] = x[i] + 1;
return ans;
}Line by line, we see the following:
The
#include<Rcpp.h> is similar tolibrary(...)in R, it brings in all that we need to write C++ code for Rcpp.using namespace Rcpp is somewhat similar todetach(...). This simplifies syntax. If we don’t include this, all calls to Rcpp members need to be explicit, e.g., instead of typingNumericVector, we would need to typeRcpp::NumericVectorThe
//starts a comment in C++, in this case, the// [[Rcpp::export]] comment is a flag Rcpp uses to “export” this C++ function to R.It is the first part of the function definition. We are creating a function that returns a
NumericVector , is calledadd1 , has a single input element namedx that is also aNumericVector .Here, we are declaring an object called
ans , which is aNumericVector with an initial size equal to the size ofx . Notice that.size() is called a “member function” of thexobject, which is of classNumericVector.We are declaring a for-loop (three parts):
int i = 0 We declare the variablei, an integer, and initialize it at 0.i < x.size() This loop will end wheni’s value is at or above the length ofx.++i At each iteration,iwill increment in one unit.
ans[i] = x[i] + 1 set the i-th element ofansequal to the i-th element ofxplus 1.return ans exists the function returning the vectorans.
Now, where to execute/run this?
- You can use the
sourceCppfunction from theRcpppackage to run .cpp scripts (this is what I do most of the time). - There’s also
cppFunction, which allows compiling a single function. - Write an R package that works with Rcpp.
For now, let’s use the first option.
7.10 Example running .cpp file
Imagine that we have the following file named norm.cpp
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
double normRcpp(NumericVector x) {
return sqrt(sum(pow(x, 2.0)));
}We can compile and obtain this function using this line Rcpp::sourceCpp("norm.cpp"). Once compiled, a function called normRcpp will be available in the current R session.
7.11 Your turn
7.11.1 Problem 1: Adding vectors
- Using what you have just learned about Rcpp, write a function to add two vectors of the same length. Use the following template
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector add_vectors([declare vector 1], [declare vector 2]) {
... magick ...
return [something];
}- Now, we have to check for lengths. Use the
stopfunction to make sure lengths match. Add the following lines in your code
if ([some condition])
stop("an arbitrary error message :)");7.11.2 Problem 2: Fibonacci series
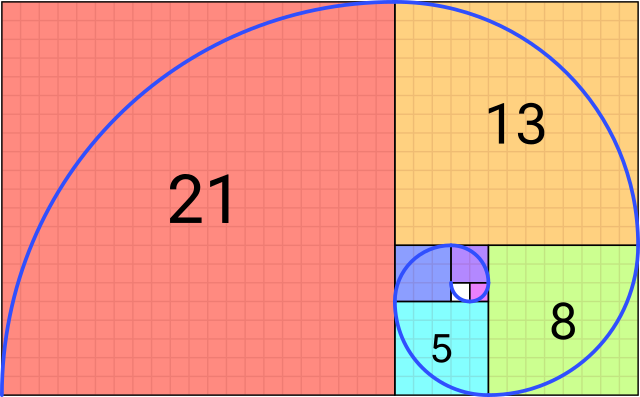
Each element of the sequence is determined by the following:
Using recursions, we can implement this algorithm in R as follows:
fibR <- function(n) {
if (n <= 1)
return(n)
fibR(n - 1) + fibR(n - 2)
}
# Is it working?
c(
fibR(0), fibR(1), fibR(2),
fibR(3), fibR(4), fibR(5),
fibR(6)
)[1] 0 1 1 2 3 5 8Now, let’s translate this code into Rcpp and see how much speed boost we get!
7.11.3 Problem 2: Fibonacci series (solution)
Code
#include <Rcpp.h>
// [[Rcpp::export]]
int fibCpp(int n) {
if (n <= 1)
return n;
return fibCpp(n - 1) + fibCpp(n - 2);
}microbenchmark::microbenchmark(fibR(20), fibCpp(20))Unit: microseconds
expr min lq mean median uq max neval
fibR(20) 6160.142 6242.056 6501.54652 6271.395 6375.465 8404.680 100
fibCpp(20) 15.770 15.995 28.28494 16.631 18.274 1044.358 1007.12 RcppArmadillo and OpenMP
Friendlier than RcppParallel… at least for ‘I-use-Rcpp-but-don’t-actually-know-much-about-C++’ users (like myself!).
Must run only ‘Thread-safe’ calls, so calling R within parallel blocks can cause problems (almost all the time).
Use
armaobjects, e.g.arma::mat,arma::vec, etc. Or, if you are used to themstd::vectorobjects as these are thread-safe.Pseudo Random Number Generation is not very straightforward… But C++11 has a nice set of functions that can be used together with OpenMP
Need to think about how processors work, cache memory, etc. Otherwise, you could get into trouble… if your code is slower when run in parallel, then you probably are facing false sharing
If R crashes… try running R with a debugger (see Section 4.3 in Writing R extensions):
~$ R --debugger=valgrind
7.12.1 RcppArmadillo and OpenMP workflow
Tell Rcpp that you need to include that in the compiler:
#include <omp.h> // [[Rcpp::plugins(openmp)]]Within your function, set the number of cores, e.g
// Setting the cores omp_set_num_threads(cores);Tell the compiler that you’ll be running a block in parallel with OpenMP
#pragma omp [directives] [options] { ...your neat parallel code... }You’ll need to specify how OMP should handle the data:
shared: Default, all threads access the same copy.private: Each thread has its own copy, uninitialized.firstprivateEach thread has its own copy, initialized.lastprivateEach thread has its own copy. The last value used is returned.
Setting
default(none)is a good practice.Compile!
7.12.2 Ex 5: RcppArmadillo + OpenMP
Our own version of the dist function… but in parallel!
#include <omp.h>
#include <RcppArmadillo.h>
// [[Rcpp::depends(RcppArmadillo)]]
// [[Rcpp::plugins(openmp)]]
using namespace Rcpp;
// [[Rcpp::export]]
arma::mat dist_par(const arma::mat & X, int cores = 1) {
// Some constants
int N = (int) X.n_rows;
int K = (int) X.n_cols;
// Output
arma::mat D(N,N);
D.zeros(); // Filling with zeros
// Setting the cores
omp_set_num_threads(cores);
#pragma omp parallel for shared(D, N, K, X) default(none)
for (int i=0; i<N; ++i)
for (int j=0; j<i; ++j) {
for (int k=0; k<K; k++)
D.at(i,j) += pow(X.at(i,k) - X.at(j,k), 2.0);
// Computing square root
D.at(i,j) = sqrt(D.at(i,j));
D.at(j,i) = D.at(i,j);
}
// My nice distance matrix
return D;
}# Simulating data
set.seed(1231)
K <- 5000
n <- 500
x <- matrix(rnorm(n*K), ncol=K)
# Are we getting the same?
table(as.matrix(dist(x)) - dist_par(x, 4)) # Only zeros
0
250000 # Benchmarking!
microbenchmark::microbenchmark(
dist(x), # stats::dist
dist_par(x, cores = 1), # 1 core
dist_par(x, cores = 2), # 2 cores
dist_par(x, cores = 4), # 4 cores
times = 1,
unit = "ms"
)Unit: milliseconds
expr min lq mean median uq
dist(x) 1280.8688 1280.8688 1280.8688 1280.8688 1280.8688
dist_par(x, cores = 1) 1089.8426 1089.8426 1089.8426 1089.8426 1089.8426
dist_par(x, cores = 2) 716.9385 716.9385 716.9385 716.9385 716.9385
dist_par(x, cores = 4) 645.0109 645.0109 645.0109 645.0109 645.0109
max neval
1280.8688 1
1089.8426 1
716.9385 1
645.0109 17.12.3 Ex 6: The future
future is an R package that was designed “to provide a very simple and uniform way of evaluating R expressions asynchronously using various resources available to the user.”
futureclass objects are either resolved or unresolved.If queried, Resolved values are return immediately, and Unresolved values will block the process (i.e. wait) until it is resolved.
Futures can be parallel/serial, in a single (local or remote) computer, or a cluster of them.
Let’s see a brief example
library(future)
plan(multicore)
# We are creating a global variable
a <- 2
# Creating the futures has only the overhead (setup) time
system.time({
x1 %<-% {Sys.sleep(3);a^2}
x2 %<-% {Sys.sleep(3);a^3}
})
## user system elapsed
## 0.031 0.017 0.049
# Let's just wait 5 seconds to make sure all the cores have returned
Sys.sleep(3)
system.time({
print(x1)
print(x2)
})
## [1] 4
## [1] 8
## user system elapsed
## 0.004 0.000 0.0047.12.4 Bonus track 1: Simulating
We know that
So, we approximate
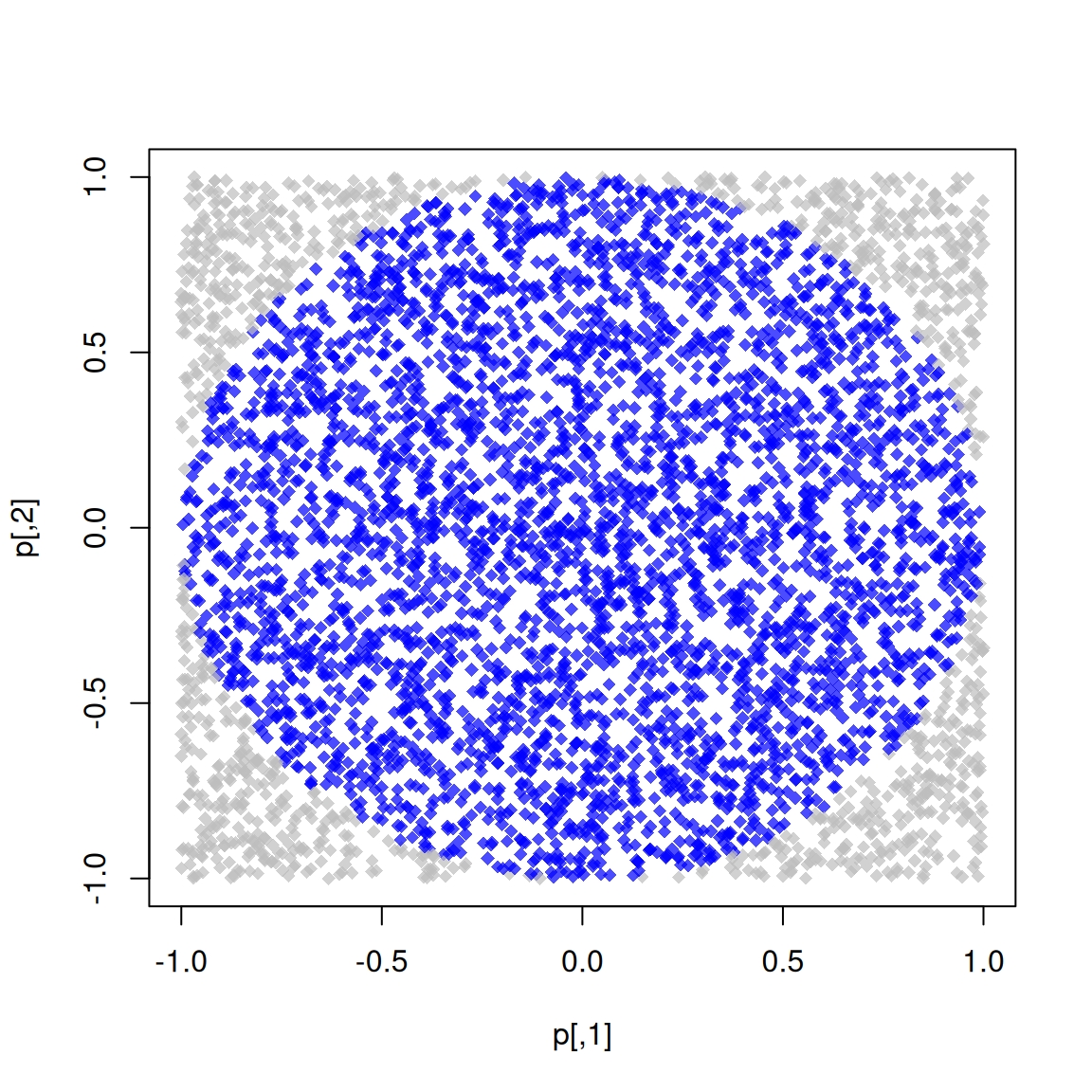
The R code to do this
pisim <- function(i, nsim) { # Notice we don't use the -i-
# Random points
ans <- matrix(runif(nsim*2), ncol=2)
# Distance to the origin
ans <- sqrt(rowSums(ans^2))
# Estimated pi
(sum(ans <= 1)*4)/nsim
}library(parallel)
# Setup
cl <- makePSOCKcluster(4L)
clusterSetRNGStream(cl, 123)
# Number of simulations we want each time to run
nsim <- 1e5
# We need to make -nsim- and -pisim- available to the
# cluster
clusterExport(cl, c("nsim", "pisim"))
# Benchmarking: parSapply and sapply will run this simulation
# a hundred times each, so at the end we have 1e5*100 points
# to approximate pi
microbenchmark::microbenchmark(
parallel = parSapply(cl, 1:100, pisim, nsim=nsim),
serial = sapply(1:100, pisim, nsim=nsim),
times = 1
)Unit: milliseconds
expr min lq mean median uq max neval
parallel 284.9262 284.9262 284.9262 284.9262 284.9262 284.9262 1
serial 379.6148 379.6148 379.6148 379.6148 379.6148 379.6148 1ans_par <- parSapply(cl, 1:100, pisim, nsim=nsim)
ans_ser <- sapply(1:100, pisim, nsim=nsim)
stopCluster(cl) par ser R
3.141762 3.141266 3.141593 7.13 See also
- Package parallel
- Using the iterators package
- Using the foreach package
- 32 OpenMP traps for C++ developers
- The OpenMP API specification for parallel programming
- ‘openmp’ tag in Rcpp gallery
- OpenMP tutorials and articles
For more, check out the CRAN Task View on HPC

